Product Information
Product Name | Product Number | Specification |
HRbio™ Ⅲ 1st Strand cDNA Synthesis Kit (OneStep gDNA Removal) One-step genomic DNA removal reverse transcription kit | HRF0191 | 10 T |
HRF0192 | 100 T |
Product Description
This product uses RNA as a template and adopts pre-mixing technology to efficiently synthesize the first-strand cDNA with 5×HRbio™ III RT Reaction Mix (pre-mixed with HRbio™ III Reverse Transcriptase, RNase Inhibitor, dNTP Mixture, Buffer), which is easy to operate.
This product uses molecular evolution technology and a new high-temperature reverse transcriptase with a temperature of up to 60°C. It can read through RNA templates with rich GC content and complex secondary structures, greatly improving the efficiency and length of reverse transcription. It can be used for longer cDNA synthesis and the construction of a high-proportion full-length cDNA library. This kit uses a special gDNA Remover with DNA decomposition activity. With just one step, genome removal and reverse transcription reactions can be completed simultaneously, greatly simplifying the operation steps and avoiding the risk of sample contamination and RNA degradation caused by complex sample addition processes.
Product Usage
First-strand cDNA synthesis. It can be used for cloning and detection of high-copy and low-copy genes, and is especially suitable for high-temperature reverse transcription of complex templates with high GC content.
Features
1. The new generation of high temperature reverse transcriptase greatly improves the efficiency and length of reverse transcription including complex RNA templates. The length of synthesized cDNA is up to 15 kb or more.
2. Fully premixed reverse transcription Mix, just add RNA, primers and water to complete reverse transcription simply and quickly.
3. The volume of RNA template can be added up to 75% of the total volume, which is very suitable for reverse transcription reactions of low-concentration RNA templates.
4. The premixed mix does not freeze at -20°C, which reduces the thawing and mixing time and makes it easier to use. Users can flexibly choose Oligo (dT), Random primer or gene-specific primer as the reverse transcription primer according to their needs.
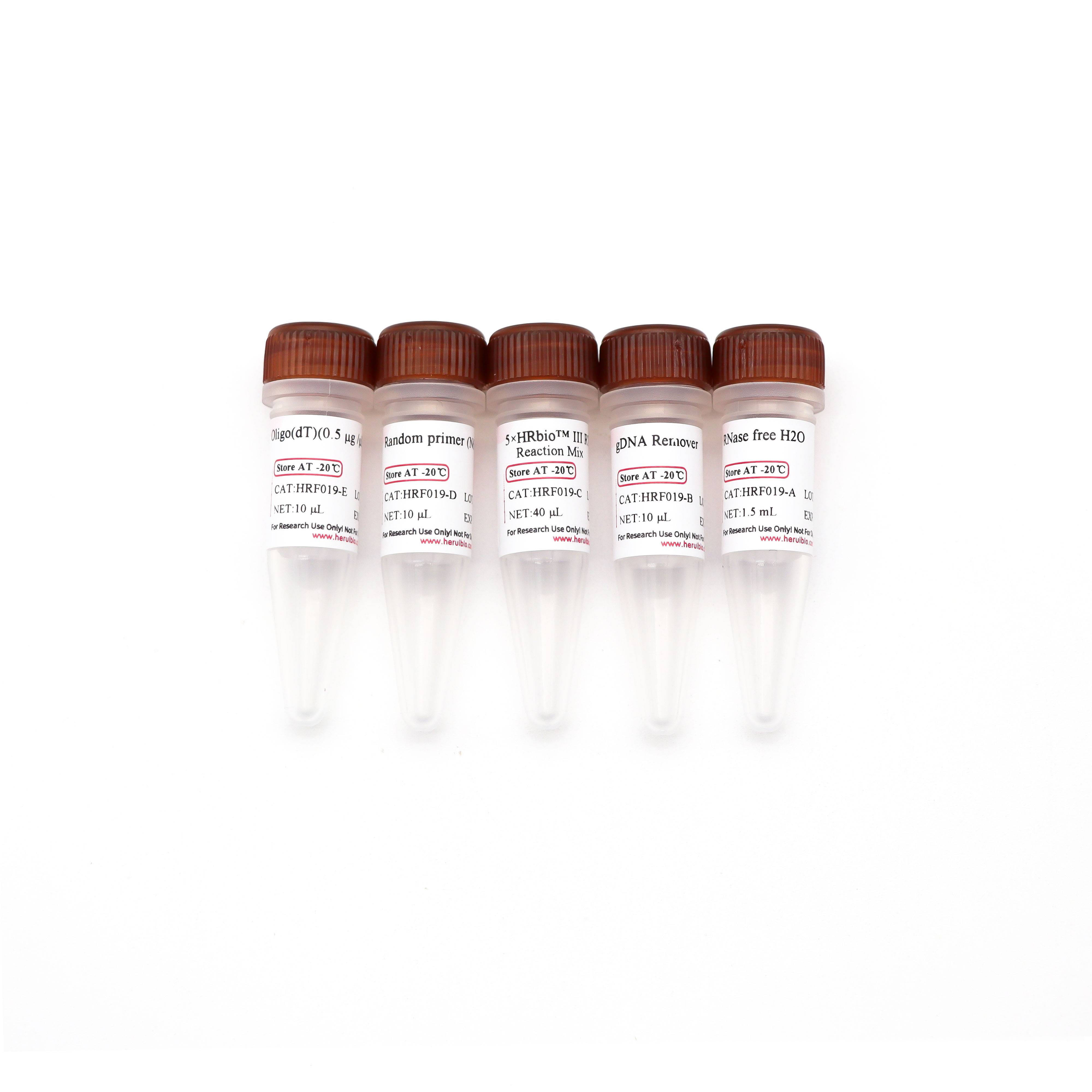
Product composition
serial number | Components | HRF0191 (10T) | HRF0192 (100T) |
HRF019-A | RNase free H2O | 1 mL | 1.5 mL |
HRF019-B | gDNA Remover | 10 μL | 100 μL |
HRF019-C | 5×HRbio™ III RT Reaction Mix | 40 μL | 400 μL |
HRF019-D | Random primer (N6) | 10 μL | 100 μL |
HRF019-E | Oligo(dT)(0.5 μg/μl) | 10 μL | 100 μL |
Transportation and storage methods
Transported by dry ice. Store at -20℃, avoid repeated freezing and thawing, valid for 1 year.
Precautions
1) Avoid RNase contamination.
2) To ensure successful reverse transcription, it is recommended to use high-quality RNA samples.
3) Optional step (usually not required) : If the RNA template is GC-rich or has a complex secondary structure, or the length of the amplified cDNA exceeds 3kb, you can first add only the RNA template, primers and RNase free H2O, mix well, denature at 65°C for 5 minutes, cool on ice, centrifuge briefly, and then add other components to continue the reverse transcription step below.
4) 5×HRbio™ III RT Reaction Mix and gDNA Remover contain glycerol, which is very viscous. The solution is easily adsorbed to the tube wall and the outside of the pipette tip, resulting in loss. Please centrifuge before use and avoid loss due to adhesion to the outer wall of the pipette tip. The enzyme contained in 5×HRbio™ III RT Reaction Mix is in excess. Even if 3.6 μl-3.8 μl is used each time, and 0.8 μl-0.9 μl of gDNA Remover is used each time, it will not affect the effect.
How to use
Primer selection:
1. If the template is from eukaryotic origin, Oligo (dT) is generally preferred. It can be paired with the 3' Poly A tail of eukaryotic mRNA to obtain the highest yield of full-length cDNA.
2. If for some species, it is uncertain whether the mRNA has a polyA tail, Oligo (dT) is the first choice. If unsuccessful, try gene-specific primers (GSP) and random primers.
3. gDNA Remover uses advanced one-step genome removal technology to complete the genomic DNA removal step in the shortest time and easiest way.
4. Gene-specific primers (GSP) have the highest specificity. However, in some cases, the GSP used in PCR reactions cannot effectively guide the synthesis of the first-strand cDNA. Oligo (dT) or random primers can be used instead for reverse transcription.
5. Random primer has the lowest specificity. All RNA, including mRNA, rRNA, and tRNA, can be used as templates for random primers. When the target region has a complex secondary structure or a high GC content, or the template is from prokaryotes, and Oligo (dT) or gene-specific primers (GSP) cannot effectively guide cDNA synthesis, random primers can be used as primers.
6. If the synthesized cDNA is used downstream for fluorescent quantitative PCR, Oligo (dT) and Random primer can be mixed (1μl of each in 20μl reaction system) to make the cDNA synthesis efficiency of each region of mRNA the same, which helps to improve the authenticity and repeatability of the quantitative results.
First-strand cDNA synthesis (taking 20 μl reaction system as an example)
1. Add the following ingredients (before use, flick or vortex each solution to mix well. Centrifuge briefly to collect liquid at the bottom of the tube.)
Components | concentration |
Total RNA/mRNA | 50 ng-5 μg/5-500 ng |
Oligo(dT)(0.5 μg /μl) or Random Primer (0.1 μg/μl) or GSP (Gene Specific Primer, 2 pmol/μl) | 1 μL |
5×HRbio™ III RT Reaction Mix | 4 μL (see Note 4) |
gDNA Remover * | 1 μL (see Note 4) |
RNase free H 2 O | to 20 μl (make up to a total volume of 20 μl) |
*If you do not need to remove genomic DNA during reverse transcription, simply omit the gDNA Remover component.
2. Mix gently
If using Oligo(dT) or Gene Specific Primers (GSP), incubate at 50°C for 30-50 min (if the product is used for qPCR, incubate at 50°C for 15 min); if using Random Primer, incubate at 25°C for 10 min, and then at 50°C for 30-50 min (if the product is used for qPCR, incubate at 50°C for 15 min)
Note: This product has good reverse transcription stability at 42°C-60°C. If the template has a complex secondary structure or a high GC region, you can try to increase the reaction temperature to 55°C-60°C to help increase the yield.
Inactivate HRbio™ III Reverse Transcriptase by heating at 3.85°C for 5 sec.
4. The obtained cDNA product can be used for PCR reaction immediately, or stored at -20°C and used within half a year; for long-term storage, it is recommended to store at -70°C after aliquoting. Repeated freezing and thawing of cDNA should be avoided.
RT-PCR
It is recommended to use 1/10-1/5 volume (2-4 μl) of the reverse transcription product as a PCR template. If the abundance is high, the cDNA can be appropriately diluted before use.
Note:
Avoid RNase contamination.
2. To ensure successful reverse transcription, it is recommended to use high-quality RNA samples.
3. Optional step (generally not required): If the RNA template is rich in GC content or has a complex secondary structure, or the length of the amplified cDNA exceeds 3kb,
you can first add only the RNA template, primers and RNase free H 2 O and mix them well, denature at 65°C for 5 minutes, cool on ice, and add
other to continue the reverse transcription step below.
4. The 5×HRbio™ III RT Reaction Mix and gDNA Remover contain glycerol and are very viscous. The solution is easily adsorbed on the wall of the tube and the outside of the pipette tip, resulting in loss.
Please centrifuge before use and avoid loss by adhering to the outer wall of the pipette tip. The enzymes contained in the 5×HRbio™ III RT Reaction Mix are all in excess.
Even if 3.6 μl-3.8 μl is used each time, and 0.8 μl-0.9 μl of gDNA Remover is used, it will not affect the effect.This product is for scientific research purposes only!