Product Information
Product Name | Product Number | Specification |
HRbio™ qPCR SYBR Green Master Mix (No Rox) | HRF0031 | 1 mL |
HRF0032 | 5×1 mL | |
HRF0033 | 50×1 mL |
Product Description
HRbio™qPCR SYBR Green Master Mix (No Rox) is a premixed solution for 2× real-time quantitative PCR amplification. The mix contains hot-start HRbio™ DNA Polymerase, SYBR Green I, dNTPs, and Mg2+. When used, you only need to add templates and primers to the amplification system to perform real-time fluorescence quantitative PCR, which greatly simplifies the operation process and reduces the chance of contamination.
The DNA polymerase ligand used in this product can regulate the activity of DNA polymerase in real time with temperature changes. The formula adds factors that effectively inhibit nonspecific PCR amplification and factors that improve the amplification efficiency of PCR reactions, so that quantitative PCR can obtain a good linear relationship in a wide quantitative range.
Applicable models
Bio-Rad: CFX96, CFX384, iCycler iQ, iQ5, MyiQ, MiniOpticon, Opticon, Opticon 2, Chromo4;
Eppendorf: Mastercycler ep realplex, realplex 2 s;
Qiagen: Corbett Rotor-Gene Q, Rotor-Gene 3000, Rotor-Gene 6000;
Roche Applied Science: LightCycler 480, LightCycler 2.0; Lightcycler 96;
Thermo Scientific: PikoReal Cycler; Cepheid: SmartCycler; Illumina: Eco qPCR.
Transportation and storage methods
Transported with ice packs. Store at -20℃, valid for 18 months.
Avoid repeated freezing and thawing of this product. The product contains the fluorescent dye SYBR Green I, so avoid strong light exposure when storing or preparing the reaction system.
Precautions
1) We recommend using our cDNA synthesis kit to effectively remove the residual genome in RNA samples.
2) After thawing, the Master Mix may have flocculent material. Place it at 4°C and mix it upside down until the solution is clear. This will not affect the performance of the reagent.
3) For your safety and health, please wear a lab coat and disposable gloves when operating.
4) This product is for scientific research purposes only!
Reaction system (preparation on ice is recommended)
Components | Volume (μL) | Volume (μL) | Final concentration |
HRbio™qPCR SYBR Green Master Mix (No Rox) | 25 | 10 | 1× |
Forward Primer (10 μM) | 1 | 0.4 | 0.2 μM |
Reverse Primer (10 μM) | 1 | 0.4 | 0.2 μM |
Template DNA | X | X | - |
Sterile ultrapure water | To 50 | To 20 | - |
【Note】: Be sure to mix thoroughly before use to avoid violent shaking to produce excessive bubbles.
a) Primer concentration: The final primer concentration is usually 0.2 μM, and can be adjusted between 0.1-1.0 μM depending on the situation.
b) Template concentration: If the template type is undiluted cDNA stock solution, the volume used should not exceed 1/10 of the total qPCR reaction volume.
c) Template dilution: The cDNA stock solution is recommended to be diluted 5-10 times. The optimal amount of template added is the amount that the CT value obtained after amplification is 20-30 cycles.
d) Reaction system: 20 μL or 50 μL is recommended to ensure the effectiveness and repeatability of target gene amplification.
e) System preparation: Please prepare in a clean bench and use gun tips and reaction tubes without nuclease residues; gun tips with filters are recommended to avoid cross contamination and aerosol contamination.
Amplification procedure (two-step method)
Cycle steps | temperature | time | Number of cycles |
Pre-denaturation | 95℃ | 5 min | 1 |
transsexual | 95℃ | 10 sec | 40 |
Annealing/Extension | 60℃ | 30 sec★ | |
Melting curve stage | Instrument Default Settings 1 | ||
Amplification procedure (three-step method)
Cycle steps | temperature | time | Number of cycles |
Pre-denaturation | 95℃ | 5 min | 1 |
transsexual | 95℃ | 10 sec | 40 |
annealing | 55-60℃ | 20 sec | |
extend | 72℃ | 20 sec★ | |
Melting curve stage | Instrument Default Settings 1 | ||
【Note】For high specificity, the two-step method can be selected, and for high efficiency amplification, the three-step method can be selected.
a) Pre-denaturation time: It can be shortened to 2 min depending on the specific conditions of different templates and primers.
b) Annealing temperature and time: Please adjust according to the length of primers and target gene.
c) Fluorescence signal acquisition (★): Please set up the experimental procedure according to the instrument manual. The time settings for several common instruments are as follows:
30sec or more: Applied Biosystems: StepOne, StepOne Plus, 7500 Fast; Roche Applied Science: LightCycler 480; Bio-Rad: CFX96
31sec and above: Applied Biosystems: 7300
34sec and above: Applied Biosystems: 7500
d) Melting curve: In general, the instrument default program can be used.
Results Analysis
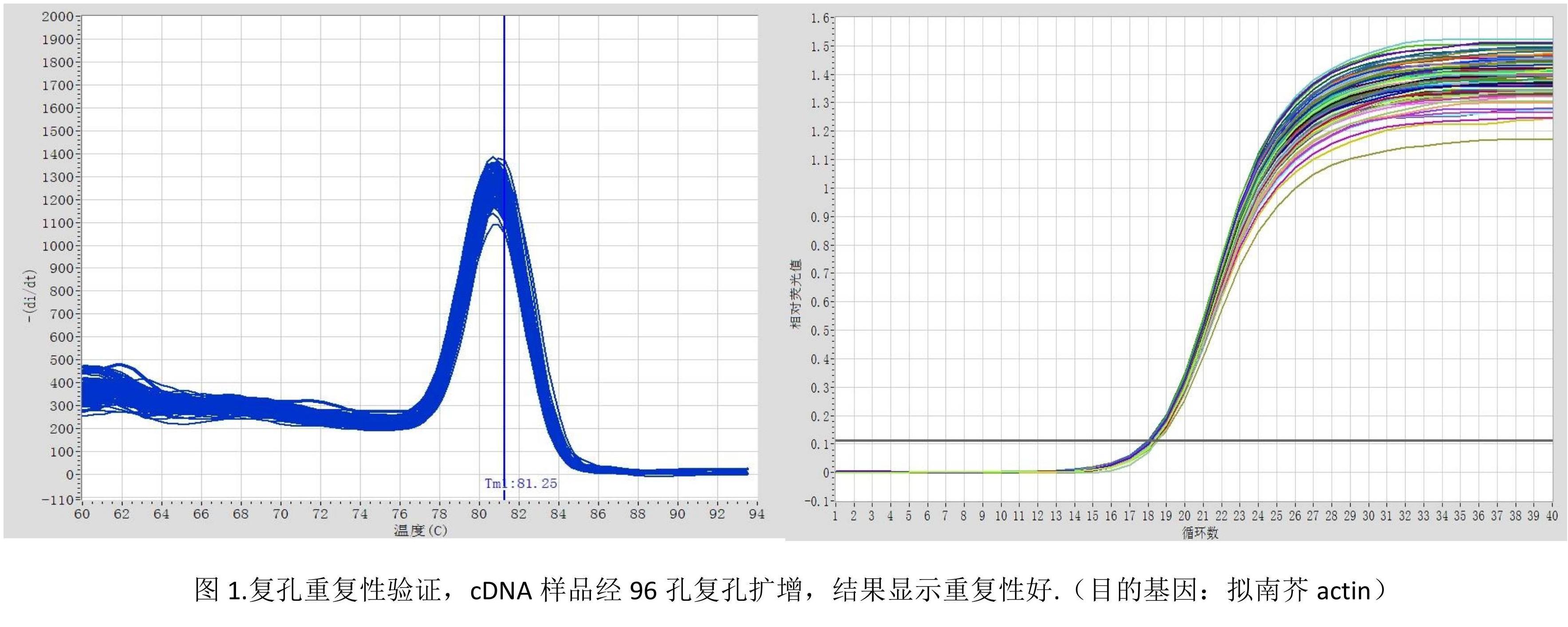
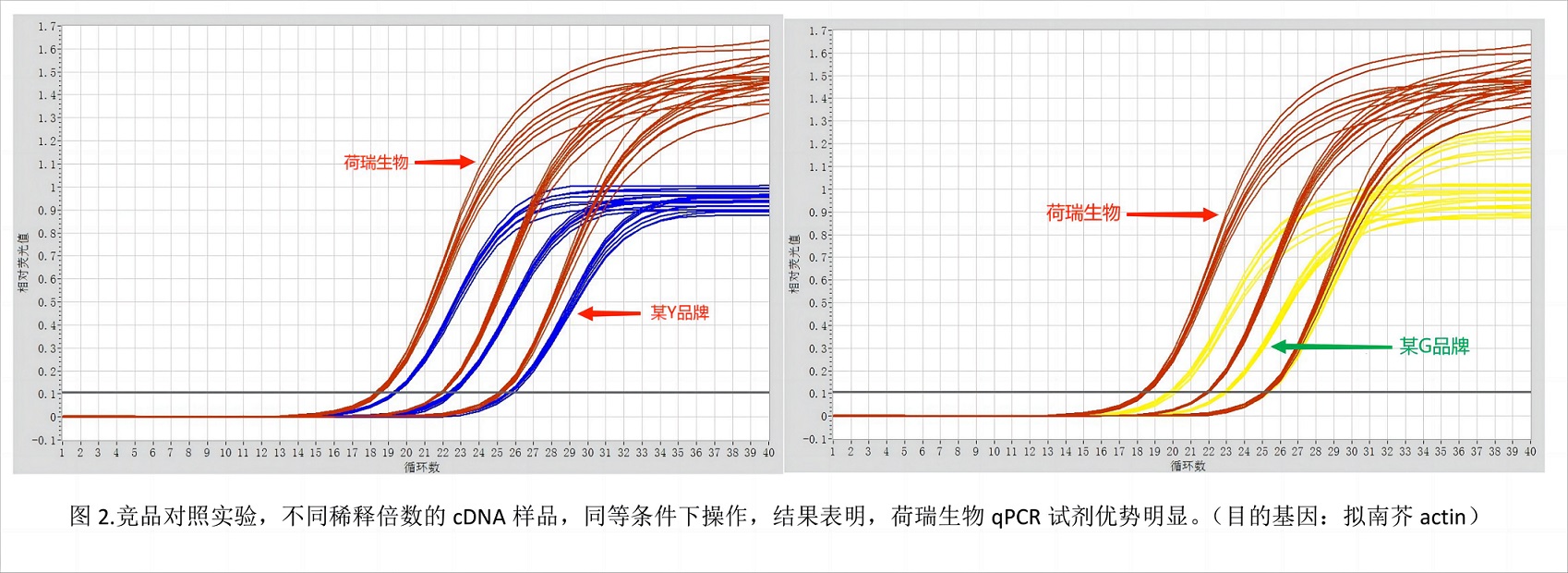
Quantitative experiments require at least three biological replicates. Amplification curves and melting curves need to be confirmed after the reaction is completed.
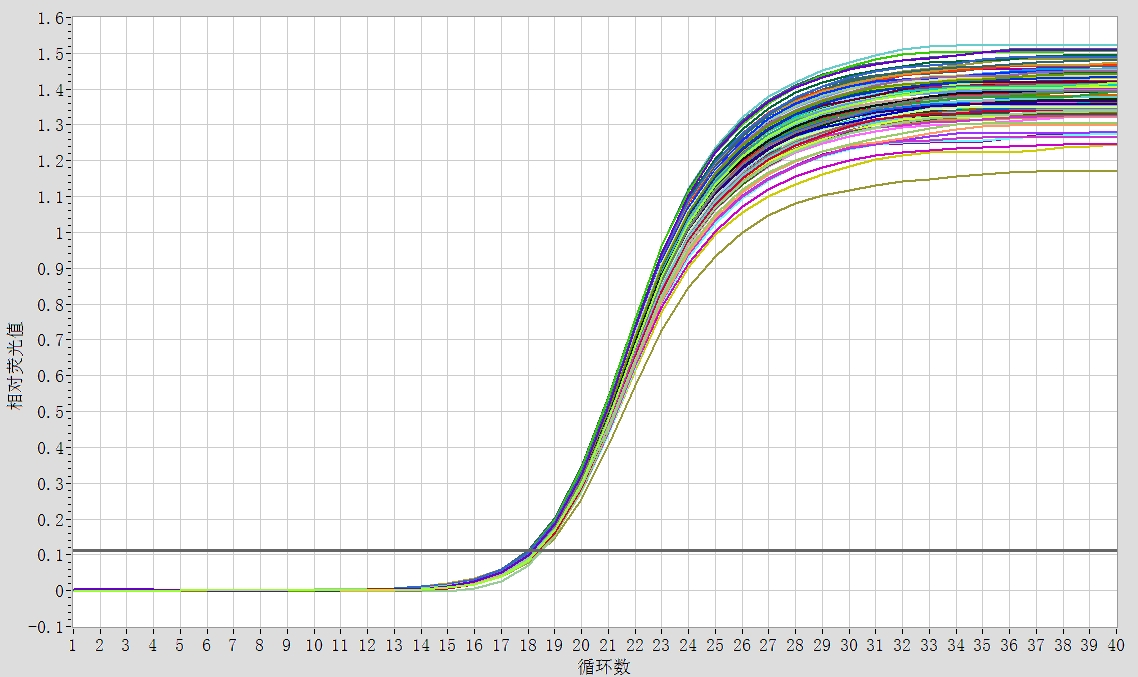
1) Amplification curve:
The standard amplification curve is S-shaped.
When the Ct value falls between 20-30, the quantitative analysis is most accurate;
If the Ct value is less than 10, the template needs to be diluted and the experiment needs to be repeated;
When the Ct value is between 30-35, it is necessary to increase the template concentration or the volume of the reaction system to improve the amplification efficiency and ensure the accuracy of the result analysis;
When the Ct value is greater than 35, the test results cannot quantitatively analyze the expression of the gene, but can be used for qualitative analysis.
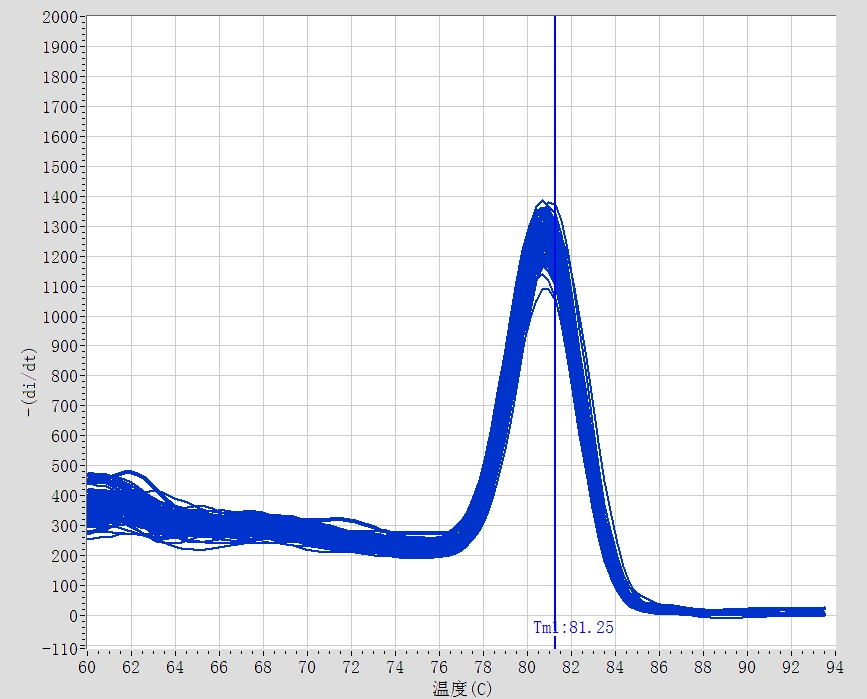
2) Melting curve:
A single peak in the melting curve indicates that the reaction specificity is good and quantitative analysis can be performed; if the melting curve has double or multiple peaks, quantitative analysis cannot be performed.
If the line shows double peaks, DNA agarose gel electrophoresis is needed to determine whether the non-target peak is primer dimer or nonspecific amplification.
If it is a primer dimer, it is recommended to reduce the primer concentration or redesign primers with high amplification efficiency.
If it is non-specific amplification, please increase the annealing temperature or redesign primers with higher specificity.
Primer Design Guide
1. The recommended primer length is about 25 bp. The optimal length of the amplified product is 150 bp, and can be selected within 100 bp-300 bp.
2. The difference between the Tm values of the forward primer and the reverse primer should not exceed 2°C. The optimal Tm value of the primer is 60°C-65°C.
3. Primer bases should be evenly distributed to avoid four consecutive identical bases, and the GC content should be controlled at around 50%. The last base at the 3' end should preferably be G or C.
4. It is best to avoid the presence of complementary sequences of more than 3 bases within the primer or between the forward and reverse primers.
5. Primer specificity needs to be checked using the NCBI BLAST program. Avoid nonspecific complementarity of more than 2 bases at the 3' end of the primer.
6. The designed primers need to be tested for amplification efficiency. Only primers with the same amplification efficiency can be used for quantitative comparative analysis.
Experimental Case